mlflow.shap
-
mlflow.shap.get_default_conda_env()[source] - Returns
The default Conda environment for MLflow Models produced by calls to
save_explainer()andlog_explainer().
-
mlflow.shap.get_default_pip_requirements()[source] A list of default pip requirements for MLflow Models produced by this flavor. Calls to
save_explainer()andlog_explainer()produce a pip environment that, at minimum, contains these requirements.
-
mlflow.shap.get_underlying_model_flavor(model)[source] Find the underlying models flavor.
- Parameters
model – underlying model of the explainer.
-
mlflow.shap.load_explainer(model_uri)[source] Load a SHAP explainer from a local file or a run.
- Parameters
model_uri –
The location, in URI format, of the MLflow model. For example:
/Users/me/path/to/local/modelrelative/path/to/local/models3://my_bucket/path/to/modelruns:/<mlflow_run_id>/run-relative/path/to/modelmodels:/<model_name>/<model_version>models:/<model_name>/<stage>
For more information about supported URI schemes, see Referencing Artifacts.
- Returns
A SHAP explainer.
-
mlflow.shap.log_explainer(explainer, artifact_path, serialize_model_using_mlflow=True, conda_env=None, code_paths=None, registered_model_name=None, signature: mlflow.models.signature.ModelSignature = None, input_example: Union[pandas.core.frame.DataFrame, numpy.ndarray, dict, list, csr_matrix, csc_matrix, str, bytes, tuple] = None, await_registration_for=300, pip_requirements=None, extra_pip_requirements=None, metadata=None)[source] Log an SHAP explainer as an MLflow artifact for the current run.
- Parameters
explainer – SHAP explainer to be saved.
artifact_path – Run-relative artifact path.
serialize_model_using_mlflow – When set to True, MLflow will extract the underlying model and serialize it as an MLmodel, otherwise it uses SHAP’s internal serialization. Defaults to True. Currently MLflow serialization is only supported for models of ‘sklearn’ or ‘pytorch’ flavors.
conda_env –
Either a dictionary representation of a Conda environment or the path to a conda environment yaml file. If provided, this describes the environment this model should be run in. At a minimum, it should specify the dependencies contained in
get_default_conda_env(). IfNone, a conda environment with pip requirements inferred bymlflow.models.infer_pip_requirements()is added to the model. If the requirement inference fails, it falls back to usingget_default_pip_requirements(). pip requirements fromconda_envare written to a piprequirements.txtfile and the full conda environment is written toconda.yaml. The following is an example dictionary representation of a conda environment:{ "name": "mlflow-env", "channels": ["conda-forge"], "dependencies": [ "python=3.8.15", { "pip": [ "shap==x.y.z" ], }, ], }
code_paths –
A list of local filesystem paths to Python file dependencies (or directories containing file dependencies). These files are prepended to the system path when the model is loaded. Files declared as dependencies for a given model should have relative imports declared from a common root path if multiple files are defined with import dependencies between them to avoid import errors when loading the model.
For a detailed explanation of
code_pathsfunctionality, recommended usage patterns and limitations, see the code_paths usage guide.registered_model_name – If given, create a model version under
registered_model_name, also creating a registered model if one with the given name does not exist.signature –
ModelSignaturedescribes model input and outputSchema. The model signature can beinferredfrom datasets with valid model input (e.g. the training dataset with target column omitted) and valid model output (e.g. model predictions generated on the training dataset), for example:from mlflow.models import infer_signature train = df.drop_column("target_label") predictions = ... # compute model predictions signature = infer_signature(train, predictions)
input_example – one or several instances of valid model input. The input example is used as a hint of what data to feed the model. It will be converted to a Pandas DataFrame and then serialized to json using the Pandas split-oriented format, or a numpy array where the example will be serialized to json by converting it to a list. Bytes are base64-encoded. When the
signatureparameter isNone, the input example is used to infer a model signature.await_registration_for – Number of seconds to wait for the model version to finish being created and is in
READYstatus. By default, the function waits for five minutes. Specify 0 or None to skip waiting.pip_requirements – Either an iterable of pip requirement strings (e.g.
["shap", "-r requirements.txt", "-c constraints.txt"]) or the string path to a pip requirements file on the local filesystem (e.g."requirements.txt"). If provided, this describes the environment this model should be run in. IfNone, a default list of requirements is inferred bymlflow.models.infer_pip_requirements()from the current software environment. If the requirement inference fails, it falls back to usingget_default_pip_requirements(). Both requirements and constraints are automatically parsed and written torequirements.txtandconstraints.txtfiles, respectively, and stored as part of the model. Requirements are also written to thepipsection of the model’s conda environment (conda.yaml) file.extra_pip_requirements –
Either an iterable of pip requirement strings (e.g.
["pandas", "-r requirements.txt", "-c constraints.txt"]) or the string path to a pip requirements file on the local filesystem (e.g."requirements.txt"). If provided, this describes additional pip requirements that are appended to a default set of pip requirements generated automatically based on the user’s current software environment. Both requirements and constraints are automatically parsed and written torequirements.txtandconstraints.txtfiles, respectively, and stored as part of the model. Requirements are also written to thepipsection of the model’s conda environment (conda.yaml) file.Warning
The following arguments can’t be specified at the same time:
conda_envpip_requirementsextra_pip_requirements
This example demonstrates how to specify pip requirements using
pip_requirementsandextra_pip_requirements.metadata – Custom metadata dictionary passed to the model and stored in the MLmodel file.
-
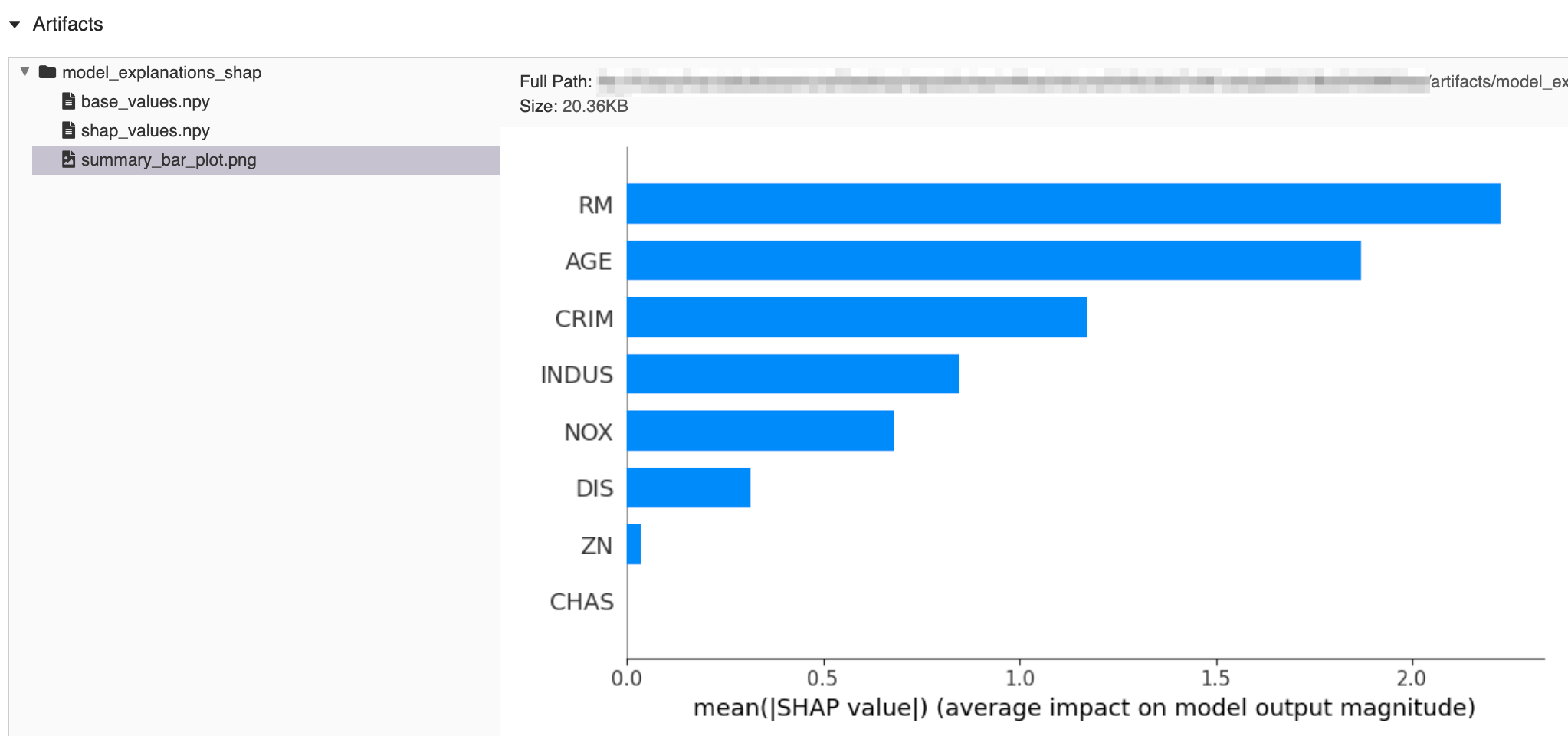
mlflow.shap.log_explanation(predict_function, features, artifact_path=None)[source] Given a
predict_functioncapable of computing ML model output on the providedfeatures, computes and logs explanations of an ML model’s output. Explanations are logged as a directory of artifacts containing the following items generated by SHAP (SHapley Additive exPlanations).Base values
SHAP values (computed using shap.KernelExplainer)
Summary bar plot (shows the average impact of each feature on model output)
- Parameters
predict_function –
A function to compute the output of a model (e.g.
predict_probamethod of scikit-learn classifiers). Must have the following signature:def predict_function(X) -> pred: ...
X: An array-like object whose shape should be (# samples, # features).pred: An array-like object whose shape should be (# samples) for a regressor or (# classes, # samples) for a classifier. For a classifier, the values inpredshould correspond to the predicted probability of each class.
Acceptable array-like object types:
numpy.arraypandas.DataFrameshap.common.DenseDatascipy.sparse matrix
features –
A matrix of features to compute SHAP values with. The provided features should have shape (# samples, # features), and can be either of the array-like object types listed above.
Note
Background data for shap.KernelExplainer is generated by subsampling
featureswith shap.kmeans. The background data size is limited to 100 rows for performance reasons.artifact_path – The run-relative artifact path to which the explanation is saved. If unspecified, defaults to “model_explanations_shap”.
- Returns
Artifact URI of the logged explanations.
import os import numpy as np import pandas as pd from sklearn.datasets import load_diabetes from sklearn.linear_model import LinearRegression import mlflow from mlflow import MlflowClient # prepare training data X, y = dataset = load_diabetes(return_X_y=True, as_frame=True) X = pd.DataFrame(dataset.data[:50, :8], columns=dataset.feature_names[:8]) y = dataset.target[:50] # train a model model = LinearRegression() model.fit(X, y) # log an explanation with mlflow.start_run() as run: mlflow.shap.log_explanation(model.predict, X) # list artifacts client = MlflowClient() artifact_path = "model_explanations_shap" artifacts = [x.path for x in client.list_artifacts(run.info.run_id, artifact_path)] print("# artifacts:") print(artifacts) # load back the logged explanation dst_path = client.download_artifacts(run.info.run_id, artifact_path) base_values = np.load(os.path.join(dst_path, "base_values.npy")) shap_values = np.load(os.path.join(dst_path, "shap_values.npy")) print("\n# base_values:") print(base_values) print("\n# shap_values:") print(shap_values[:3])
# artifacts: ['model_explanations_shap/base_values.npy', 'model_explanations_shap/shap_values.npy', 'model_explanations_shap/summary_bar_plot.png'] # base_values: 20.502000000000002 # shap_values: [[ 2.09975523 0.4746513 7.63759026 0. ] [ 2.00883109 -0.18816665 -0.14419184 0. ] [ 2.00891772 -0.18816665 -0.14419184 0. ]]
-
mlflow.shap.save_explainer(explainer, path, serialize_model_using_mlflow=True, conda_env=None, code_paths=None, mlflow_model=None, signature: mlflow.models.signature.ModelSignature = None, input_example: Union[pandas.core.frame.DataFrame, numpy.ndarray, dict, list, csr_matrix, csc_matrix, str, bytes, tuple] = None, pip_requirements=None, extra_pip_requirements=None, metadata=None)[source] Save a SHAP explainer to a path on the local file system. Produces an MLflow Model containing the following flavors:
- Parameters
explainer – SHAP explainer to be saved.
path – Local path where the explainer is to be saved.
serialize_model_using_mlflow – When set to True, MLflow will extract the underlying model and serialize it as an MLmodel, otherwise it uses SHAP’s internal serialization. Defaults to True. Currently MLflow serialization is only supported for models of ‘sklearn’ or ‘pytorch’ flavors.
conda_env –
Either a dictionary representation of a Conda environment or the path to a conda environment yaml file. If provided, this describes the environment this model should be run in. At a minimum, it should specify the dependencies contained in
get_default_conda_env(). IfNone, a conda environment with pip requirements inferred bymlflow.models.infer_pip_requirements()is added to the model. If the requirement inference fails, it falls back to usingget_default_pip_requirements(). pip requirements fromconda_envare written to a piprequirements.txtfile and the full conda environment is written toconda.yaml. The following is an example dictionary representation of a conda environment:{ "name": "mlflow-env", "channels": ["conda-forge"], "dependencies": [ "python=3.8.15", { "pip": [ "shap==x.y.z" ], }, ], }
code_paths –
A list of local filesystem paths to Python file dependencies (or directories containing file dependencies). These files are prepended to the system path when the model is loaded. Files declared as dependencies for a given model should have relative imports declared from a common root path if multiple files are defined with import dependencies between them to avoid import errors when loading the model.
For a detailed explanation of
code_pathsfunctionality, recommended usage patterns and limitations, see the code_paths usage guide.mlflow_model –
mlflow.models.Modelthis flavor is being added to.signature –
ModelSignaturedescribes model input and outputSchema. The model signature can beinferredfrom datasets with valid model input (e.g. the training dataset with target column omitted) and valid model output (e.g. model predictions generated on the training dataset), for example:from mlflow.models import infer_signature train = df.drop_column("target_label") predictions = ... # compute model predictions signature = infer_signature(train, predictions)
input_example – one or several instances of valid model input. The input example is used as a hint of what data to feed the model. It will be converted to a Pandas DataFrame and then serialized to json using the Pandas split-oriented format, or a numpy array where the example will be serialized to json by converting it to a list. Bytes are base64-encoded. When the
signatureparameter isNone, the input example is used to infer a model signature.pip_requirements – Either an iterable of pip requirement strings (e.g.
["shap", "-r requirements.txt", "-c constraints.txt"]) or the string path to a pip requirements file on the local filesystem (e.g."requirements.txt"). If provided, this describes the environment this model should be run in. IfNone, a default list of requirements is inferred bymlflow.models.infer_pip_requirements()from the current software environment. If the requirement inference fails, it falls back to usingget_default_pip_requirements(). Both requirements and constraints are automatically parsed and written torequirements.txtandconstraints.txtfiles, respectively, and stored as part of the model. Requirements are also written to thepipsection of the model’s conda environment (conda.yaml) file.extra_pip_requirements –
Either an iterable of pip requirement strings (e.g.
["pandas", "-r requirements.txt", "-c constraints.txt"]) or the string path to a pip requirements file on the local filesystem (e.g."requirements.txt"). If provided, this describes additional pip requirements that are appended to a default set of pip requirements generated automatically based on the user’s current software environment. Both requirements and constraints are automatically parsed and written torequirements.txtandconstraints.txtfiles, respectively, and stored as part of the model. Requirements are also written to thepipsection of the model’s conda environment (conda.yaml) file.Warning
The following arguments can’t be specified at the same time:
conda_envpip_requirementsextra_pip_requirements
This example demonstrates how to specify pip requirements using
pip_requirementsandextra_pip_requirements.metadata – Custom metadata dictionary passed to the model and stored in the MLmodel file.